2. Methods¶
2.1. Procedure & Visual Stimuli¶
fMRI data was collected from 61 participants.
Forty-five minutes of fMRI data was collected on every participant.
Two circles moved around randomly on the projection screen.
When circles collided, a mild but unpleasant electric shock was delivered to the index and middle fingers of participant’s left hand.
Shock delivery was meant to induce fear of circle collision.
Several “near-miss” events occured at random times during the experiment.
Near-miss events are defined as those when the circles approach each other at least for 8 seconds, come very close (i.e., distance less than 1.5 times the circle diameter), but miss and start to retreat at least for 8 seconds.
Near-miss events were included to investigate the brain’s dynamic response to approaching and retreating threats.
2.2. Data Preprocessing¶
fMRI time-series data suffers from a lot of noise such as,
Physiological signals associated with respiratory and cardiac cycles.
Head motion.
Scanner noise (drifts).
These noise were filtered out using ICA (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/).
2.3. Feature Selection¶
Time-series sequences corresponding to near-miss events were used in this project.
Each sequence was 14 timepoints long (14 x 1.25 = 17.5 seconds): first 7 timepoints corresponded to approach, and later 7 to retreat.
Every participant had 46 near-miss sequences.
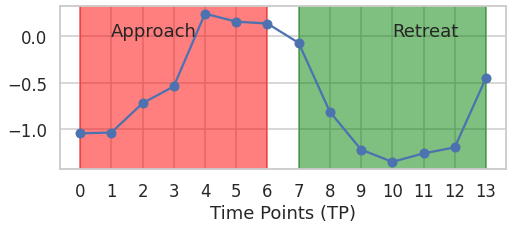
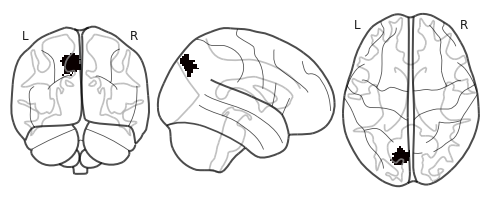
Following is an example of a sequence from the parietal cortex of the brain.
## Load the data
DATPAT = "../../data/processed/00a-ROI316_withShiftedSegments.pkl"
with open(DATPAT,"rb") as f:
SEGMENTS=pickle.load(f)
roi_idx = 108
dataset=selective_segments(SEGMENTS,5)
roi = dataset['CON054']['data'][:,roi_idx,0]
plt.figure(figsize=(8,3))
plt.plot(np.arange(14),roi,marker='o')
plt.axvspan(0,6,color='red',alpha=0.5)
plt.axvspan(7,13,color='green',alpha=0.5)
plt.text(1,0,'Approach',fontdict={'size':18})
plt.text(10,0,'Retreat',fontdict={'size':18})
#_=plt.xticks(np.arange(14)*1.25, labels=['TR{:02d}'.format(tr) for tr in np.arange(14)],rotation=90)
_=plt.xticks(np.arange(14))
_=plt.xlabel('Time Points (TP)')
mask = nil.image.load_img("../../data/processed/masks/00b-Schaefer2018_300Parcels_17Networks_order_afniMNI152_2mm_GM.nii.gz")
roi_mask = np.zeros_like(mask.get_fdata())
roi_mask[mask.get_fdata() == roi_idx] = 1.
roi_mask = nil.image.new_img_like(mask, roi_mask)
#plotting.view_img(roi_mask,symmetric_cmap=False,vmax=1)
plot_glass_brain(roi_mask)
<nilearn.plotting.displays.OrthoProjector at 0x7f1fbf8e7390>
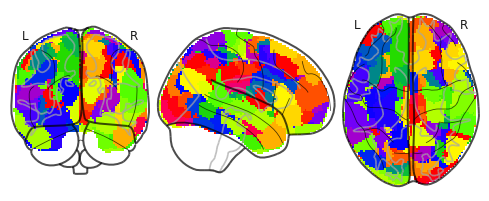
Near-miss sequences from a total of 316 brain regions were extracted in a similar fashion.
Following figure shows all the 316 brain regions of interest (ROI).
_=plot_glass_brain(mask,cmap='prism')
<nilearn.plotting.displays.OrthoProjector at 0x7f1fbdfd5780>
Each sequence was divided into two training examples and labeled accordingly as follows:
<----------- Timeseries ---------------> Label training example 1: [TP00, TP01, TP02, TP03, TP04, TP05, TP06] -> "approach" training example 2: [TP07, TP08, TP09, TP10, TP11, TP12, TP13] -> "retreat"
Out of 61 participants, only 42 participants’ data was used to train and validate the model. Remaining 19 participants data was used to test the model.
2.4. Model¶

A variant of Long-Short Term Memory (LSTM), Gated Recurrent Units (GRU) architecture was employed to characterize the spatio-temporal pattern in the fMRI data.
The GRU architecture had three hidden layers, each with 16 GRU units, and an ouput time-distributed, dense layer (DL) with single sigmoid activation unit. The time-distributed DL returned prediction at every time-point. Following was the employed model architecture:
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
gru (GRU) (None, None, 16) 16032
_________________________________________________________________
gru_1 (GRU) (None, None, 16) 1632
_________________________________________________________________
gru_2 (GRU) (None, None, 16) 1632
_________________________________________________________________
time_distributed (TimeDistri (None, None, 1) 17
=================================================================
Total params: 19,313
Trainable params: 19,313
Non-trainable params: 0
_________________________________________________________________